Documentation
- Introduction
- How to use PHARP
- Samples in PHARP
- Running time
- FAQs
- Contact us
To facilitate studies of genetic variation and their roles in the molecular mechanism of pig complex traits, we developed the Pig Haplotypes Reference Panel (PHARP, https://alphaindex.zju.edu.cn/PHARP/index.php) database, an integrated and interactive knowledge base of pig genomes and their utilization for imputation.
In the current release, PHARP provides the following:
PHARP demonstrated superior imputation accuracy compared to existing panels (SWIM, AHCpanel, AGIDB), achieving concordance rates (CR > 0.99) and correlation coefficients (R² > 0.98) in European breeds and improved accuracy in Chinese Jinhua pigs (CR = 0.936, R² = 0.924).
Overall, as a comprehensive genomic data resource with an imputation focus, PHARP provides a convenient platform for efficient phasing and imputation to facilitate genomic research of variants and their roles in pig complex traits.
In the current release, PHARP provides the following:
- 12,898 haplotypes (6,449 individuals, related to 157 pig breeds).
- 50.3 million SNPs and 5.8 million indels of autosomes.
- Web-based analytical tools to phase low-density genotypes (genotyped by commonly used commercial SNP arrays technology such as the Illumina PorcineSNP50 Genotyping BeadChip, the Illumina PorcineSNP60 v2 Genotyping BeadChip, and the GeneSeek GGP Porcine 80K SNP chip) and impute them into high-density genotypes.
Overall, as a comprehensive genomic data resource with an imputation focus, PHARP provides a convenient platform for efficient phasing and imputation to facilitate genomic research of variants and their roles in pig complex traits.
Download the variants information: PHARP_v4_sites_info.gz
Download the raw data: PHARP.bam.zip
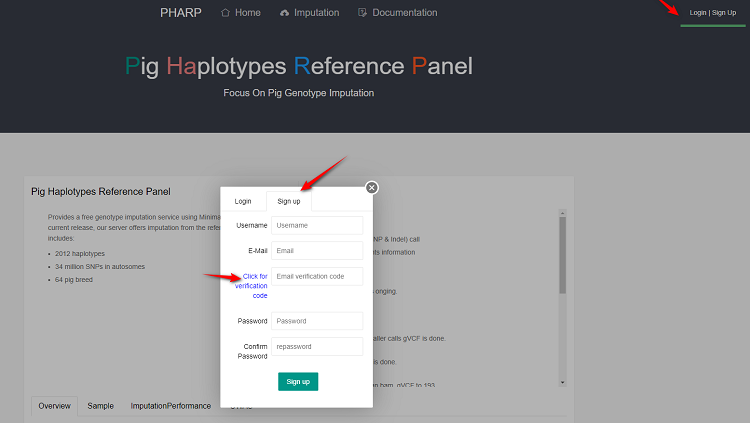
1.Sign up

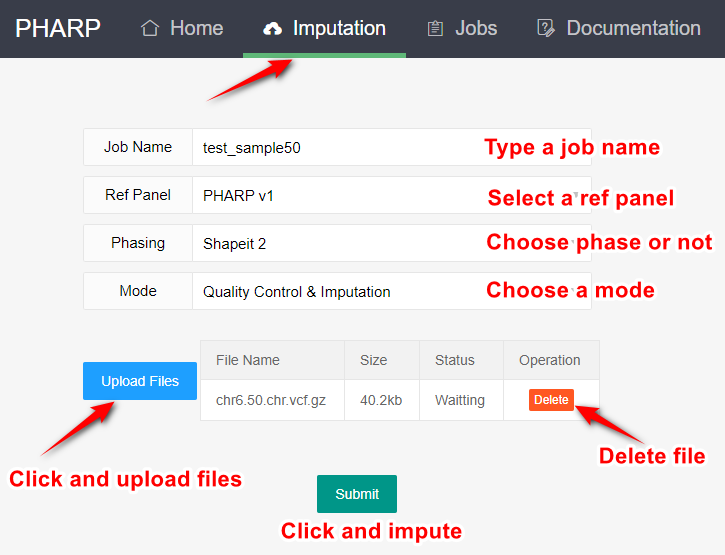
2.Upload your files for imputation
3.Check processes
4.Download your results

- Type a username and login pHARP with this later.
- Make sure your email address is correct.
2.Upload your files for imputation

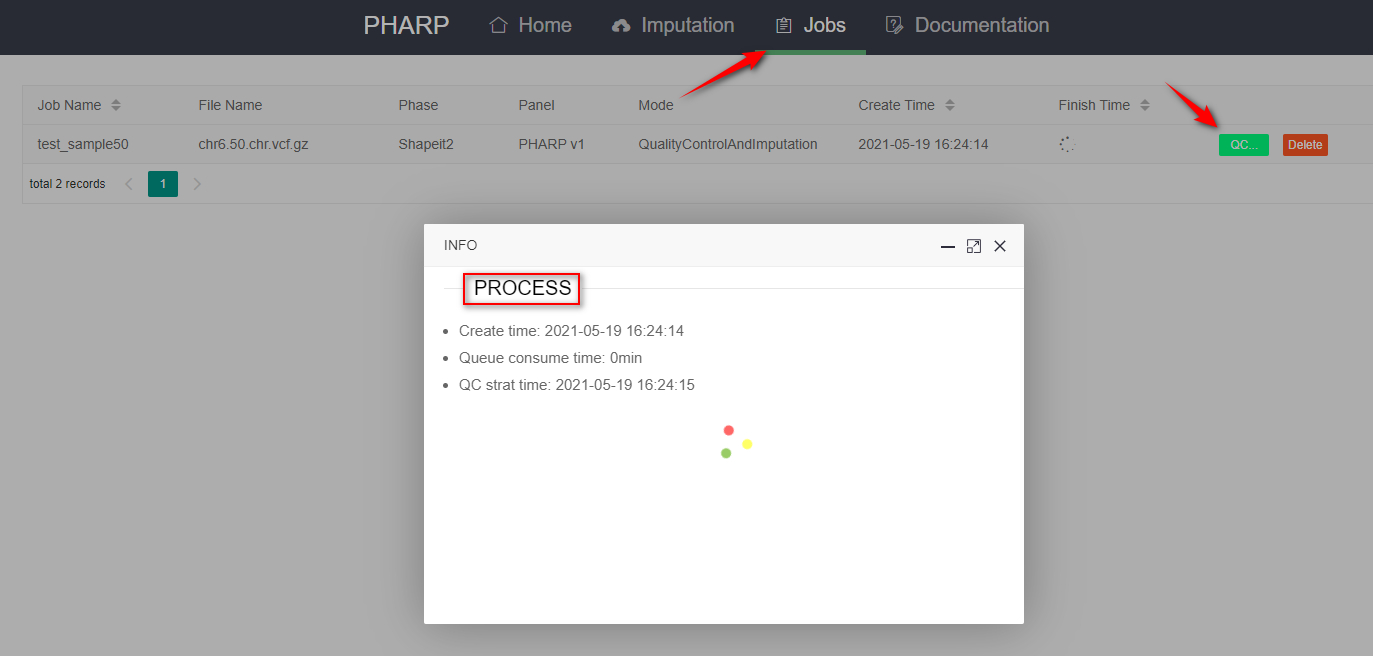
3.Check processes

- You can check your processes in the "job".
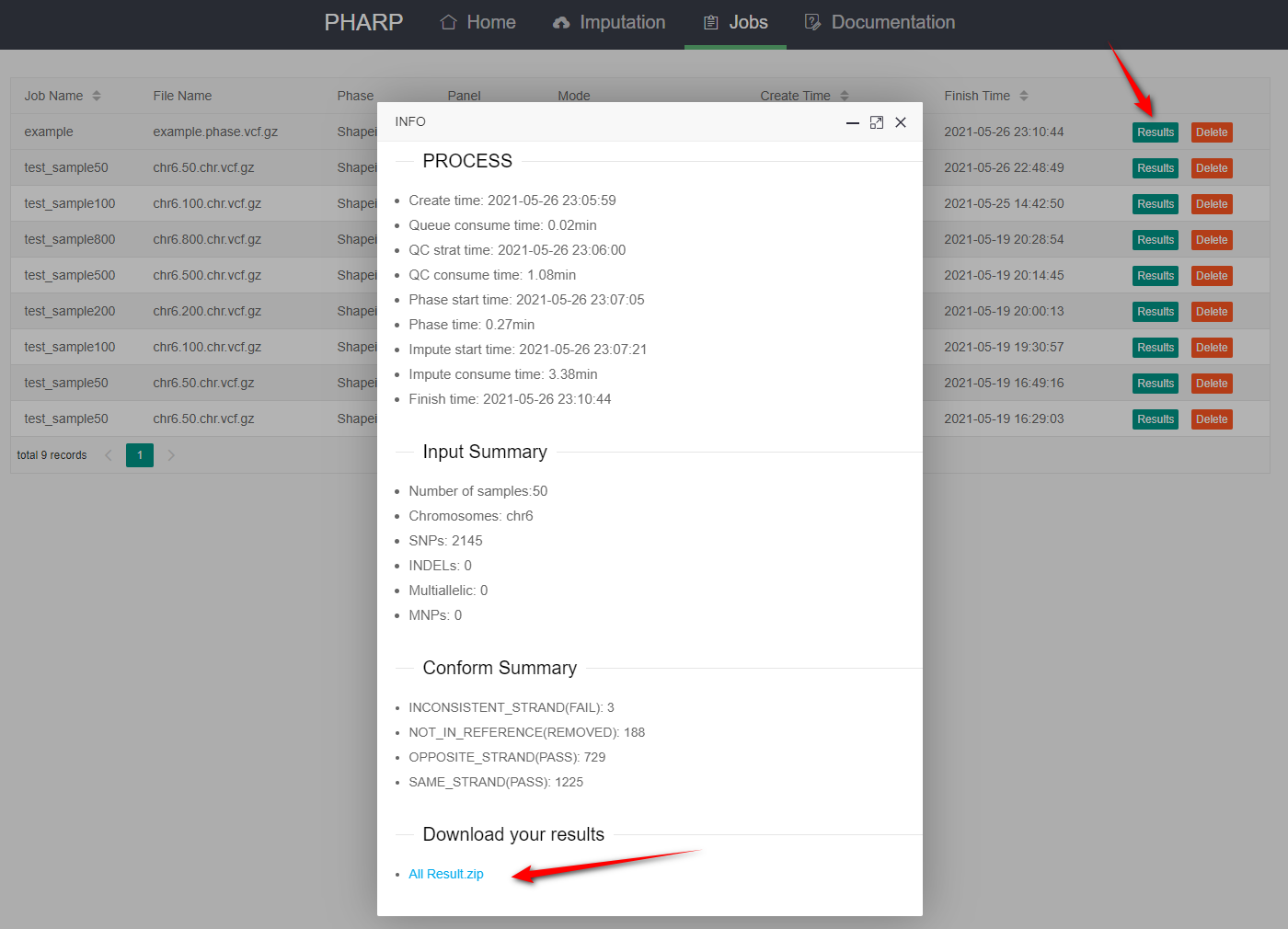
4.Download your results

- Download your results by clicking the results link.
- Meanwhile you will receive an email containing the password for unzipping your results (remember to check your email).
Abbreviations
Estimated by GGP 50K, chr6 (2,145 SNPs)
1 . Which pig assembly was used in PHARP ?
2 . How to change chromosome names?
3 . How to obtain a subset of VCF file for one chromosome from a VCF file containing multiple chromosomes ?
4 . How to convert the plink to VCF format?
5 . How to compress the VCF file?
6. Which parameter can be used for quality control of imputed-genotypes ?
PHARP assembly is based on Sscrofa11.1.
You can download it from here.
2 . How to change chromosome names?
bcftools annotate --rename-chrs chr_name.txt chr1_18.vcf.gz | bgzip -c > chr1_18.vcf.gz
# the chr_name.txt is like this 1 chr1 2 chr2 3 chr3 4 chr4 ...
3 . How to obtain a subset of VCF file for one chromosome from a VCF file containing multiple chromosomes ?
# for the vcf file
for q in {1..18}; do
vcftools --vcf chr1_18.vcf --chr chr$q --recode --out chr$q
done
# for the compressed vcf file
for q in {1..18}; do
vcftools --gzvcf chr1_18.vcf.gz --chr chr$q --recode --out chr$q
done
# for the plink format file
for q in {1..18}; do
plink --bfile chr1_18 --chr chr$q --make-bed --out chr$q
done
4 . How to convert the plink to VCF format?
plink --bfile plink --recode vcf-iid --out vcf
5 . How to compress the VCF file?
bgzip your_file.vcf
6. Which parameter can be used for quality control of imputed-genotypes ?
After getting the results, you can filter it using the 'R2' paramter for quality control.
bcftools filter -i 'R2>0.4' result.vcf.gz -Oz -o result_R2.0.4.vcf.gz
| People | |
|---|---|
| Zhenyang Zhang | zzy2014@sjtu.edu.cn |
| Zhen Wang | wangzhen20@zju.edu.cn |
| Yuchun Pan | panyc@zju.edu.cn |